Dr. Bradley Harding is a scientist specializing in DNA-based computing. His research, recently published in the prestigious journal Nano Letters, holds the potential to advance the field reusable biodevices controlled by logical operations.
Dr. Harding’s research was co-supervised by the University of the Sunshine Coast’s Dr Joanne Macdonald and Dr Nina Pollak, assisted by collaborator Professor Darko Stefanovic from the University of New Mexico.
The following is an edited transcript of my telephone conversation with Dr. Harding.
How did you get into the field?
It’s kind of a funny story. When I asked Dr. Macdonald, the professor who was my principle supervisor, about doing a PhD focused on virology—making genetically-engineered viruses—she said, “That’s a silly idea. You’ll never get a high impact factor publication by publishing in the virology journals,” which speaks volumes of the publish-or-perish nature of research. She suggested a more physics-based approach where I would be more likely to get into a high-ranking journal. She had previously worked with deoxyribozymes for DNA computing. Me being young and naïve at the time, I thought that sounded great.
How would you sum of the state of play in the field toady vs five years ago?
A lot has happened in five years. From the outside looking in, it might not seem like much has happened. But from the inside it has been a lot of small, incremental advancements that have all added up to a greater whole. We’ve made more progress in DNA-based computers in five years than what a standard silicon computer would have made within five years at its relative time of development. At that time, computers were making small advancements from vacuum tubes, moving towards a micro transistor. The DNA-based computing field has made similar advancements. We’ve gone from really basic units that can only do one thing to two and three different things. So, we are slowly progressing towards our version of a micro transistor.
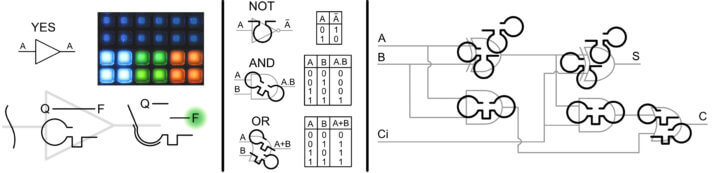
So these are bio-devices. Is that a way to describe a biological entity yoked into computation?
This is where definitions can get a little awkward. Whilst we are working with DNA, which is historically the cornerstone of biology, we use it strictly as a chemical. It’s just a chemical that we’re forcing to be in certain situations where it will energetically favor one state over another. So that allows us to say, ok, it’s taken this form therefore it’s a zero – or it’s taken the opposite form, therefore it’s a one.
How scalable is this computational property?
To a degree, it is scalable. However, the science isn’t at the point where we can produce as many gates or as many logical systems as we need. Currently we can do very simple eight-bit computations, and in the next decade we should be able scale up to 16-bit or even 32-bit.
Inside a living cell, there are all sorts of things going on. Chemical reactions, signals etc. This must all be based on computation.
To some degree in our world, everything is an expression of mathematics.
How would you describe the next step in your field or in your research?
The next step is to take the computational systems that I was working with and use them to regulate gene expression. So, instead of using a signal protein to control RNA synthesis through DNA replication it would be using a DNA control circuit, allowing a much finer-grain control over the entire system.
It sounds like you’re moving from simple instructions to conditional instructions.
Yes. That would be full Boolean logic at that point, and that’s still many years away.
The initial conditions for computation must be an ability to sense a state or some environmental condition – and that would trigger computation.
Yes, our simple logic gates were detecting the presence or absence of a second strand of DNA, our input. So, when that strand was present, we would have our logic gates turn over a substrate that would emit light. We also did more complex work that’s not published yet, where rather than detecting DNA or RNA, we could detect small molecules.
That sounds like an immunology function.
It’s funny you say that because it’s all based on aptamer technology. Aptamers have been colloquially referred to as oligonucleotide antibodies because they show the same level of specificity as an antibody would. I can’t say much more than that until the embargo on my thesis is lifted.
I thought it would be helpful if you can describe the difference between biological informatics element and the compute element you’ve been working on.
In terms of general computation – and this applies to bio-computing – there’s two actions you can undertake. You can take arithmetic operations which is the physical computing: one plus one equals two. Then there’s actuation, the outcome of the computation: if two then push, or if three then pull. But that actuation can also be the recording or reading of data. In DNA-based computers, the data storage element is not a computational element. And currently the systems are not at the point where computational elements talk to the data storage elements because they are two very different physical processes. They are biologically and chemically different.
How does this compare to CRISPR technology?
CRISPR has a few more moving parts than what we have. Our problem is stitching DNA back together. So we’re maybe ten years out before we can start using DNA as RAM. Plus CRISPR has the problem of imprecision.
One limiting factor in DNA-based computing is reuse.
Yes, this problem has been struggled with quite a bit. It’s an energetically-favorable chemical reaction – though it’s a one-off reaction. It’s the same as if you were to take magnesium metal and put it into water. The water and the magnesium turn into hydrogen gas and magnesium oxide with a lot of heat emission. You can’t recover it, at least not without some fancy chemistry. It’s the same with DNA-based computing. Once you set it off, the reaction proceeds to completion.
So reversing means going back to a naïve state?
Yes. This is one of the first things I worked on. This is where the circuit started before the addition of input. The goal was to go back to that naïve state. We got pretty close to that with the deoxyribozymes, which are the enzymatic DNA particles.
Why use DNA? Why not some other molecule?
It is the single most well understood molecule, where we know its energy structure and its chemical structure, that we can manipulate in such a way to predictably change these structures. And we have ready access to DNA. There is no chemical on earth that is studied better.
Another entity that is in a naïve state is stem cells. Do you believe they have potential to be used for DNA computing applications?
My capacity to answer is limited and likely to annoy some people. I think stem cells are a massive piece of kit with the potential to create some truly magnificent systems. I mean making all the biological systems as computational systems. So, you can go from having a single stem cell to having a complete autonomous system capable of some really complex tasks.
Does quorum sensing play any role in your research?
It would be quite interesting to see if we could take the principle of quorum sensing and large-scale diffusion and chemical reactions and see if we can incorporate that into a DNA-based computing system of some kind. There is likely some kind of network effects, perhaps using light or luminescence in a LIFI system. It’s very futuristic.
Does bio-computing have any potential in liquid biopsy technologies?
Yes, when you’ve got potentially thousands of different targets that you need to be hitting simultaneously to detect the presence of disease, bio-computing could come into a light of its own, where you would be able to assay everything present. You would be able to detect different diseases by looking at patterns of matching proteins or types of DNA. I could see DNA-based computing taking a really big presence in this field over the next five to ten years. Also, DNA-based computers can be deployed easily, perhaps without even the need for an external power source.
How are startups and the VC world getting involved with biological computing?
There are bits and pieces here and there. You don’t normally hear about startups focusing entirely on bio-computing. They normally do bio-computing as ancillary project within a company. In these cases, they may be focusing on taking a whole cell instead of just molecules like DNA. In five years, there will be more companies seeking to focus on wastewater treatment, bio-remediation, or medical diagnostics. Maybe even medical treatment.
Is there anything you want to add that I didn’t cover?
When we’re taking about DNA-based computing, or even bio-computing, we’re not talking about something that is living. Instead, this is something that is algorithmic. It is something much larger than we realize it to be. It has the capacity to surpass what a living thing can do – in theory it can surpass even what a human brain can do. It’s only limitation is what we perceive it can do. The potential is that we go from a single cell and scale it up in a very short period of time to solve a problem, such as an oil spill cleanup.

